Welcome to GAPeDNA !
The app is designed to browse taxonomic coverage of online genetic database for designated metabarcoding primers, and download the sequences.
Taxa covered include freshwater and marine fish at the moment.
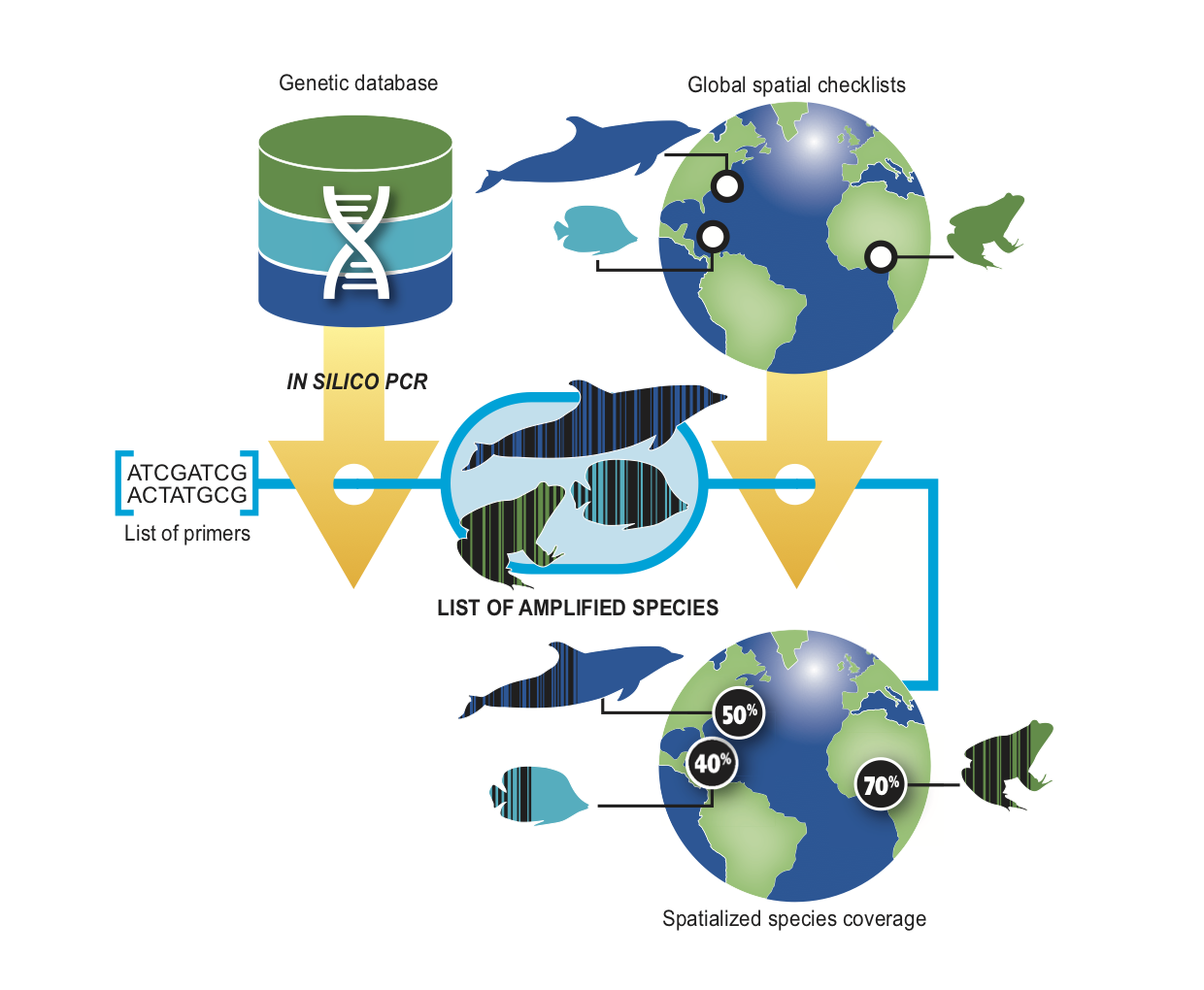
Here is how the data is generated:
Important:
Sequences in online databases must have the primer sequences, otherwise they will fail to be in-silico PCR amplified.
This might lead to under-estimated taxonomic coverage if many sequences are present but lack primers. Sequences are amplified if primer sequences have up to 3 mismatches maximum, for each primer.
Contributions to GAPeDNA:
We can expand the taxonomic breath covered by GAPeDNA with your help.
If you have spatialized checklists and/or a list of metabarcoding primer pairs for additional taxonomic groups freely available, please contact me, I can update the app and include it.
E-mail: virginie[.]marques[at]cefe.cnrs.fr
Checklist source data:
- Marine fish:
The marine fish foodweb is globally connected
- Freshwater fish:
A global database on freshwater fish species occurrence in drainage basins
Fish name were verified using the fishbase taxonomy
References
Developer and maintainer: Virginie Marques
Illustration: P. Lopez
Shiny server maintenance (CEFE): M.Q. Quidoz
GAPeDNA supports the following paper:
GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding
Source code
